CRISPR Gene Knockout Stable Cell Line
VectorBuilder는 GOI를 영구적으로 knockout시킨 안정적인 세포주를 제작할 수 있습니다. 안정적인 knockout은 CRISPR 기반 방법을 사용하여 진행됩니다. GOI를 타겟으로 하는 단일 gRNA 또는 한쌍의 gRNA를 Cas9와 함께 세포에 도입하여 DSB (double strand breaks)를 생성합니다. 이러한 breaks의 수선은 일반적으로 frameshift mutation 또는 fragment deletions이 발생하여 표적 유전자의 기능이 상실됩니다.
중점 사항
- Full expertise in CRISPR:10년 이상의 in vitro 및 in vivo 유전자 편집 경험을 보유한 전문가 팀이 성공적인 유전자 knockout을 달성하기 위해 최적의 유전자 타겟팅 전략을 디자인할 수 있습니다.
- Non-viral delivery approach: electroporation-based RNP delivery는 효율적이고 안전하며 off-target effect가 낮습니다.
- Rapid turnaround: knockout 세포를 빠르면 9주 만에 제작할 수 있습니다.
서비스 상세
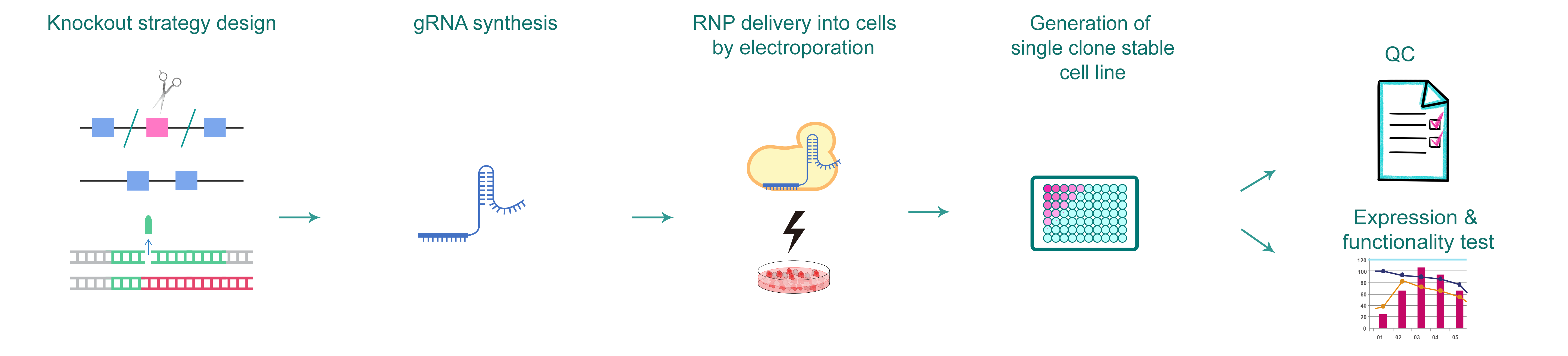
Figure 1.CRISPR로 유전자 knockout 세포를 제작하는 일반적인 워크플로우.
가격 및 소요시간
| Service Type | Deliverable | Price (USD) * | Turnaround |
|---|---|---|---|
| Knockout (frameshift mutation) | Two homozygous single clones (>10 6cells/vial, 2 vials per clone) | From $3,999 | 6-12 weeks |
| Knockout (deletion mutation) | One homozygous single clone (>10 6cells/vial, 2 vials) | From $5,999 | 6-12 weeks |
* Additional charge will apply for extra single clones or vials.
QC assays
| Assay | Methods |
|---|---|
| Knockout validation (default) | PCR, Sanger sequencing |
| Expression tests (add-on) | RT-qPCR, WB, IF, FACS |
| Off-target analysis (add-on) | NGS, PCR, Sanger sequencing |
| Chromosome analysis (add-on) | Karyotyping |
| Sterility (default) | PCR for mycoplasma detection, bioburden test for sterility detection |
다운스트림 서비스
VectorBuilder는 proliferation, apoptosis, migration, viability, cytotoxicity 등에 대한 분석을 포함하여 제작된 세포주의 다양한 phenotype 평가 및 기능 검증을 수행할 수 있습니다.
사례 연구
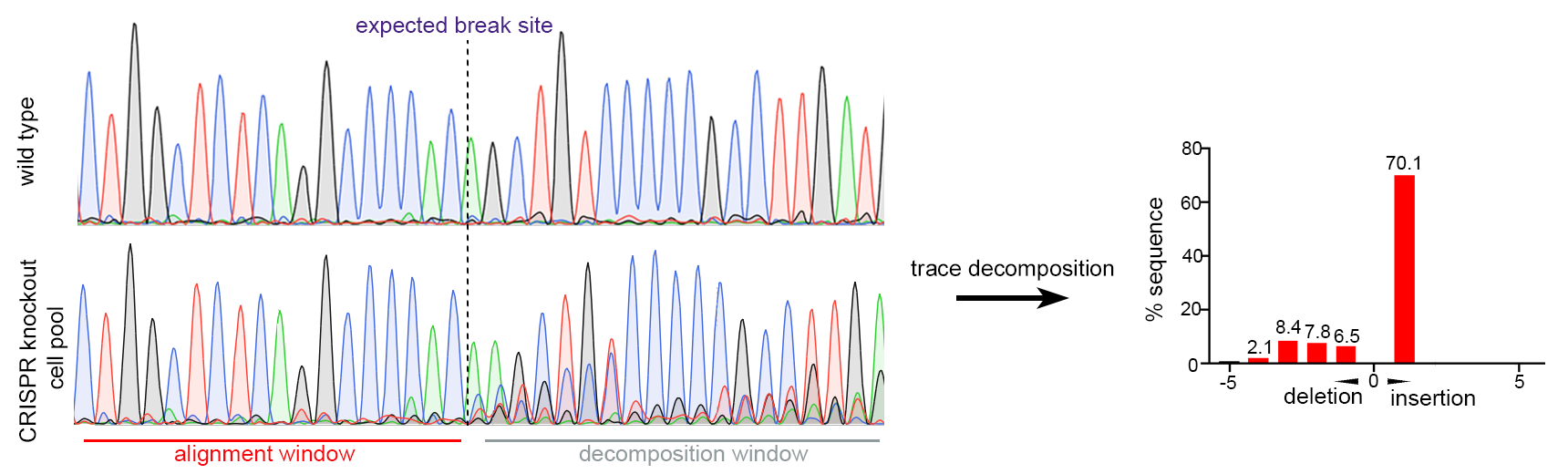
Figure 2. Huh-7 세포에서 Single gRNA CRISPR-mediated gene knockout. Pooled cells의 Sanger 시퀀싱은 CRISPR 타겟 사이트 주변에 효율적인 돌연변이를 보여주었습니다 (97.6% of sequence with mutation inferred by trace decomposition).
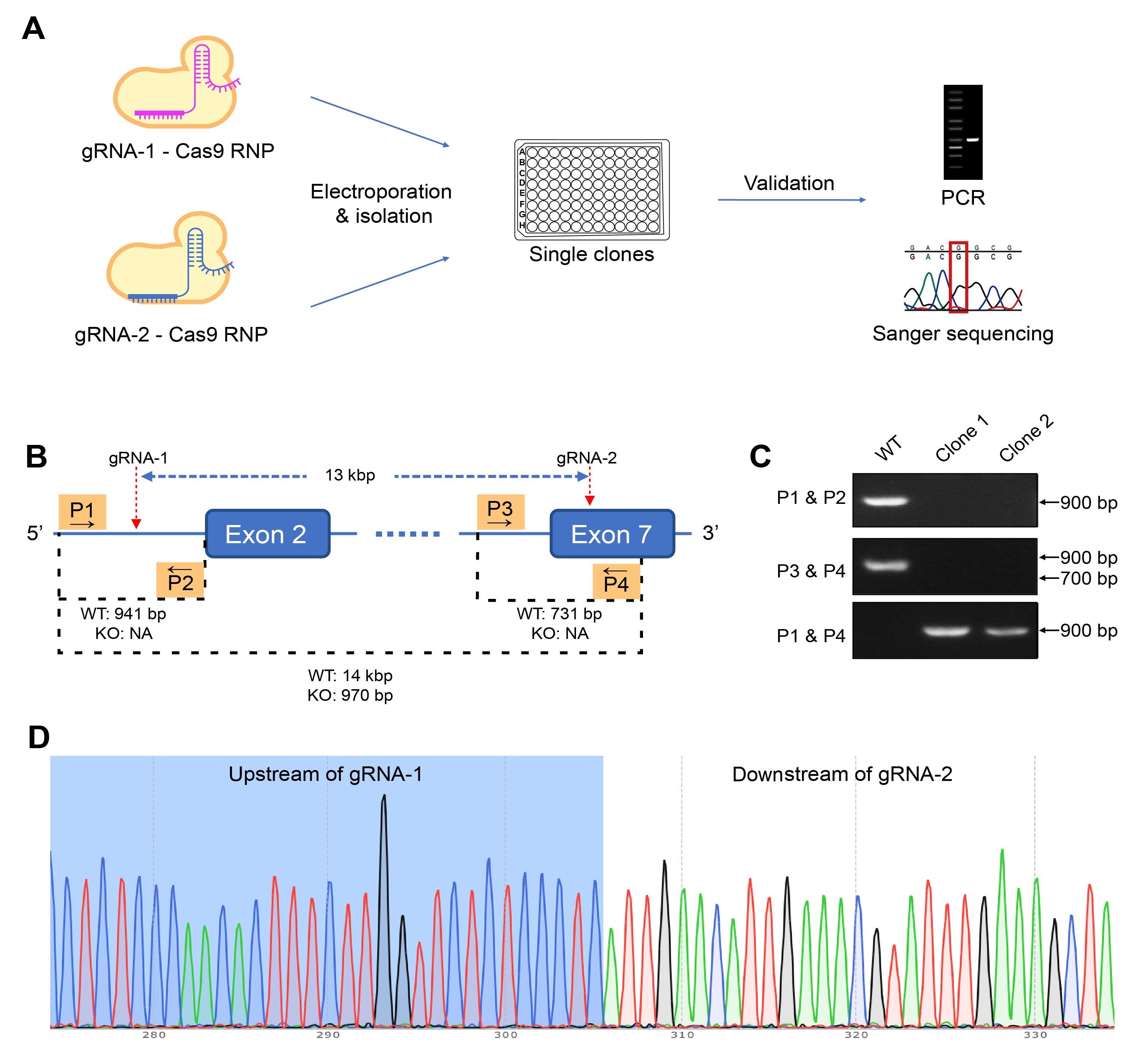
Figure 3. gRNA-Cas9 RNP (ribonucleoprotein) 방법을 사용하여 homozygous CD274 knockout (KO) 돌연변이 생성. (A) 편집 RNP를 타겟 세포에 electroporation하고, 단일 클론을 분리하여 스크리닝하였습니다. 후보군의 genotype은 PCR 및 Sanger 시퀀싱을 사용하여 검증하였습니다. (B) Murine colon adenocarcinoma cell line을 편집하는 이 사례 연구에서, 13 kbp 영역을 KO하는 타켓 유전자의 두 부위에 결합하는 RNP로 세포에 electroporation하였습니다. KO 및 WT 클론을 구별하기 위해 4개의 primer (P1~P4)를 3개의 PCR에 사용했습니다. (C) PCR 결과를 바탕으로 클론 1은 homozygous KO mutants인 것으로 검증되었으며, 이는 (D) 시퀀싱 결과에서도 확인되었습니다.
주문 방법
자료
FAQ
Which strategy to employ will depend largely on the target cell and gene. Frameshift mutations using a single gRNA are often sufficient to produce a nonfunctional gene. However, for genes with high levels of compensation, knocking out fragments of multiple genes may be necessary through fragment knockout.
In addition, dual gRNAs can be used if Cas9 nickase is being used to target the two opposite strands of a single target site. In this approach, the nickase enzyme will generate single strand cuts on both strands, one guided by each of the two gRNAs, resulting in DSBs at the target site. Generally, this method reduces off-target effects of CRISPR/Cas9 expression because targeting by both gRNAs is necessary for DSBs to be generated.
In order to decide which method is optimal for your specific application, there are a few things you should consider.
Mechnisms
- Knockdown vectors
Knockdown vectors express short hairpin RNAs (shRNAs) that repress the function of target mRNAs within the cell by inducing their cleavage and repressing their translation. Therefore, shRNA knockdown vectors are not associated with any DNA level sequence change of the gene of interest.
- Knockout vectors
CRISPR functions by directing the Cas9 nuclease to cut specific target sites in the genome. These cuts are then inefficiently repaired by the cellular machinery, resulting in permanent mutations, such as small insertions or deletions, at the sites of repair. A subset of these mutations will result in loss of function of the gene of interest due to frameshifts, premature stop codons, etc. If two closely positioned cut sites in the genome (i.e. within several kb) are targeted simultaneously, this can also result in the deletion of the intervening region.
Effectiveness
shRNA-mediated knockdown will never completely repress the expression of the target gene. Even for the most effective shRNAs, some residual expression of the target gene will remain. In contrast, in a fraction of treated cells, CRISPR can generate permanent mutations which may result in complete loss of gene function.
Consistency and uniformity
shRNA vectors generally provide high cell-to-cell uniformity within the pool of treated cells and very consistent results between experiments. In contrast, CRISPR produces results that are highly non-uniform from cell to cell due to the stochastic nature of the mutations introduced. To fully knock out the GOI in a cell, all copies of the gene in the cell must be knocked out. Given that normal cells have two copies of any gene (except for X- or Y-linked genes) while cancer cells can have more than two copies, such full knockout cells may represent a very small fraction of all the treated cells. For this reason, CRISPR knockout experiments require the screening of clones by sequencing to identify the subset in which all copies of the GOI have been knocked out.
Off-target effects
Off-target effects have been reported for both shRNA-mediated knockdown and CRISPR-mediated knockout. The off-target phenotype(s) can be estimated by using multiple different shRNAs to target the same gene. If a gene knocked down by multiple different shRNAs results in consistent phenotype(s), then it argues against the phenotype(s) being caused by off-target effects. For CRISPR-mediated knockout, multiple clones containing loss-of-function mutations can be analyzed in order to account for any phenotype(s) that may be due to off-target mutations. Additionally, bioinformatically identified off-target sites could be sequenced in the clones to see if they have been mutated.