Gene Knockdown Stable Cell Line
VectorBuilder는 GOI의 장기간 억제가 필요한 응용 분야를 위해 shRNA knockdown 안정 세포주를 제작할 수 있습니다. GOI의 효율적인 knockdown을 보장하기 위해 GOI를 타겟으로 디자인한 3개의 shRNA 중 하나를 운반하는 렌티바이러스로 표적 세포를 각각 개별적으로 transduction합니다. RT-qPCR을 통해 검증된 최고의 knockdown 효율을 보이는 세포주가 제공됩니다. 또한, 최종 세포주 제품을 출시하기 전에 무균 테스트, mycoplasma 검출 등 일련의 표준 QC 분석을 수행합니다.
중점 사항
- Permanent integration: 렌티바이러스 transduction은 호스트 세포 및 모든 자손의 게놈으로 안정적인 통합을 허용합니다.
- Broad tropism: VSV-G pseudotyped 렌티바이러스는 대부분의 포유류 세포의 표면 수용체에 결합할 수 있어 타겟 세포의 효율적인 transduction을 촉진합니다.
- Efficient down-regulation: 3가지 knockdown shRNA 중 가장 효과적인 것이 사용됩니다.
- Rapid turnaround: knockdown 세포는 벡터 디자인부터 빠르면 8주 만에 제작할 수 있습니다.
서비스 상세
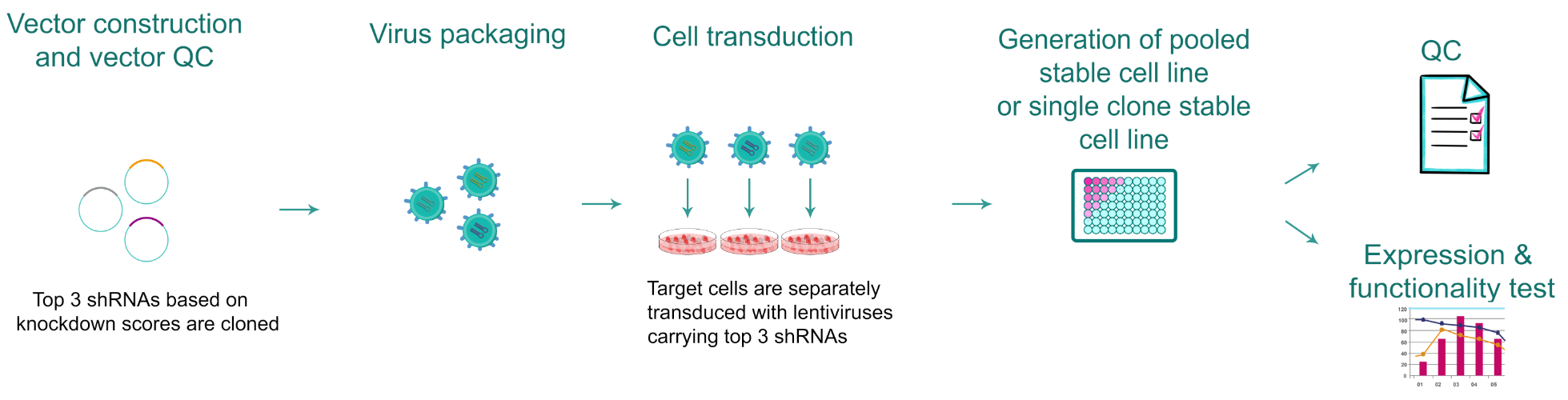
Figure 1. Typical workflow of gene knockdown stable cell line production.
가격 및 소요시간
| Service Type | Deliverable | Price (USD)* | Turnaround |
|---|---|---|---|
| Gene knockdown | Mixed pool (>106 cells/vial, 2 vials) | From $3,999 | 8-13 weeks |
| Two single clones (>106 cells/vial, 2 vials per clone) | From $4,999 | 13-18 weeks |
* Additional charge will apply for extra single clones or vials.
QC assays
| Assays | Methods |
|---|---|
| Knockdown validation (default) | RT-qPCR |
| Expression test (add-on) | WB, IF, FACS |
| Sterility (default) | PCR for mycoplasma detection, bioburden test for sterility detection |
다운스트림 서비스
VectorBuilder는 proliferation, apoptosis, migration, viability, cytotoxicity 등에 대한 분석을 포함하여 제작된 세포주의 다양한 phenotype 평가 및 기능 검증을 수행할 수 있습니다.
사례 연구
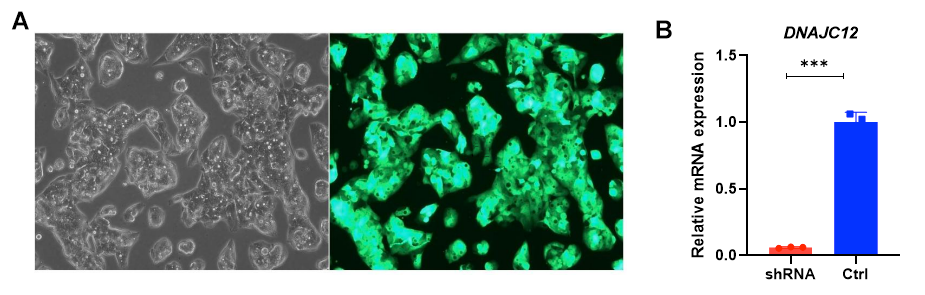
Figure 2. High DNAJC12 knockdown efficiency achieved in Hep-G2 cells. (A) Images of Hep-G2 cells transduced with lentivirus carrying DNAJC12 shRNA with EGFP marker. (B) The knockdown efficiency was 90.9% determined by RT-qPCR. Results were obtained by three repetitions; p value <0.001.
주문 방법
FAQ
VectorBuilder applies rules similar to that used by the RNAi consortium (TRC) to design and score shRNAs. For each given RefSeq transcript, we search for all possible 21mers that are considered as candidate target sites. Candidates are excluded if they contain features thought to reduce knockdown efficiency/specificity or clonability, including a run of ≥4 of the same base, a run of ≥7 G or C, GC content <25% or>60%, and AA at the 5’ end. Knockdown scores are penalized for candidates that contain internal stem-loop, high GC content toward the 3’ end, known miRNA seed sequences, or off-target matches to other genes. For genes with alternative transcripts, target sites that exist in all transcripts are given higher scores.
All scores are ≥0, with mean at ~5, standard deviation at ~5, and 95% of scores ≤15. An shRNA with a knockdown score of about 15 is considered to have the best knockdown performance and clonability, while an shRNA with a knockdown score of 0 has the worst knockdown performance or is hard to be cloned.
Please note that knockdown scores are only a rough guide. Actual knockdown efficiency could depart significantly from what the scores predict. Target sites with low scores may still work well. Also, please note that targeting 3’ UTR can be as effective as targeting coding region.
In order to decide which method is optimal for your specific application, there are a few things you should consider.
Mechanisms
- Knockdown vectors
Knockdown vectors express short hairpin RNAs (shRNAs) that repress the function of target mRNAs within the cell by inducing their cleavage and repressing their translation. Therefore, shRNA knockdown vectors are not associated with any DNA level sequence change of the gene of interest.
- Knockout vectors
CRISPR functions by directing the Cas9 nuclease to cut specific target sites in the genome. These cuts are then inefficiently repaired by the cellular machinery, resulting in permanent mutations, such as small insertions or deletions, at the sites of repair. A subset of these mutations will result in loss of function of the gene of interest due to frameshifts, premature stop codons, etc. If two closely positioned cut sites in the genome (i.e. within several kb) are targeted simultaneously, this can also result in the deletion of the intervening region.
Effectiveness
shRNA-mediated knockdown will never completely repress the expression of the target gene. Even for the most effective shRNAs, some residual expression of the target gene will remain. In contrast, in a fraction of treated cells, CRISPR can generate permanent mutations which may result in complete loss of gene function.
Consistency and uniformity
shRNA vectors generally provide high cell-to-cell uniformity within the pool of treated cells and very consistent results between experiments. In contrast, CRISPR produces results that are highly non-uniform from cell to cell due to the stochastic nature of the mutations introduced. To fully knock out the GOI in a cell, all copies of the gene in the cell must be knocked out. Given that normal cells have two copies of any gene (except for X- or Y-linked genes) while cancer cells can have more than two copies, such full knockout cells may represent a very small fraction of all the treated cells. For this reason, CRISPR knockout experiments require the screening of clones by sequencing to identify the subset in which all copies of the GOI have been knocked out.
Off-target effects
Off-target effects have been reported for both shRNA-mediated knockdown and nuclease-mediated knockout. The off-target phenotype(s) can be estimated by using multiple different shRNAs to target the same gene. If a gene knocked down by multiple different shRNAs results in consistent phenotype(s), then it argues against the phenotype(s) being caused by off-target effects. For CRISPR-mediated knockout, multiple clones containing loss-of-function mutations can be analyzed in order to account for any phenotype(s) that may be due to off-target mutations. Additionally, bioinformatically identified off-target sites could be sequenced in the clones to see if they have been mutated.



