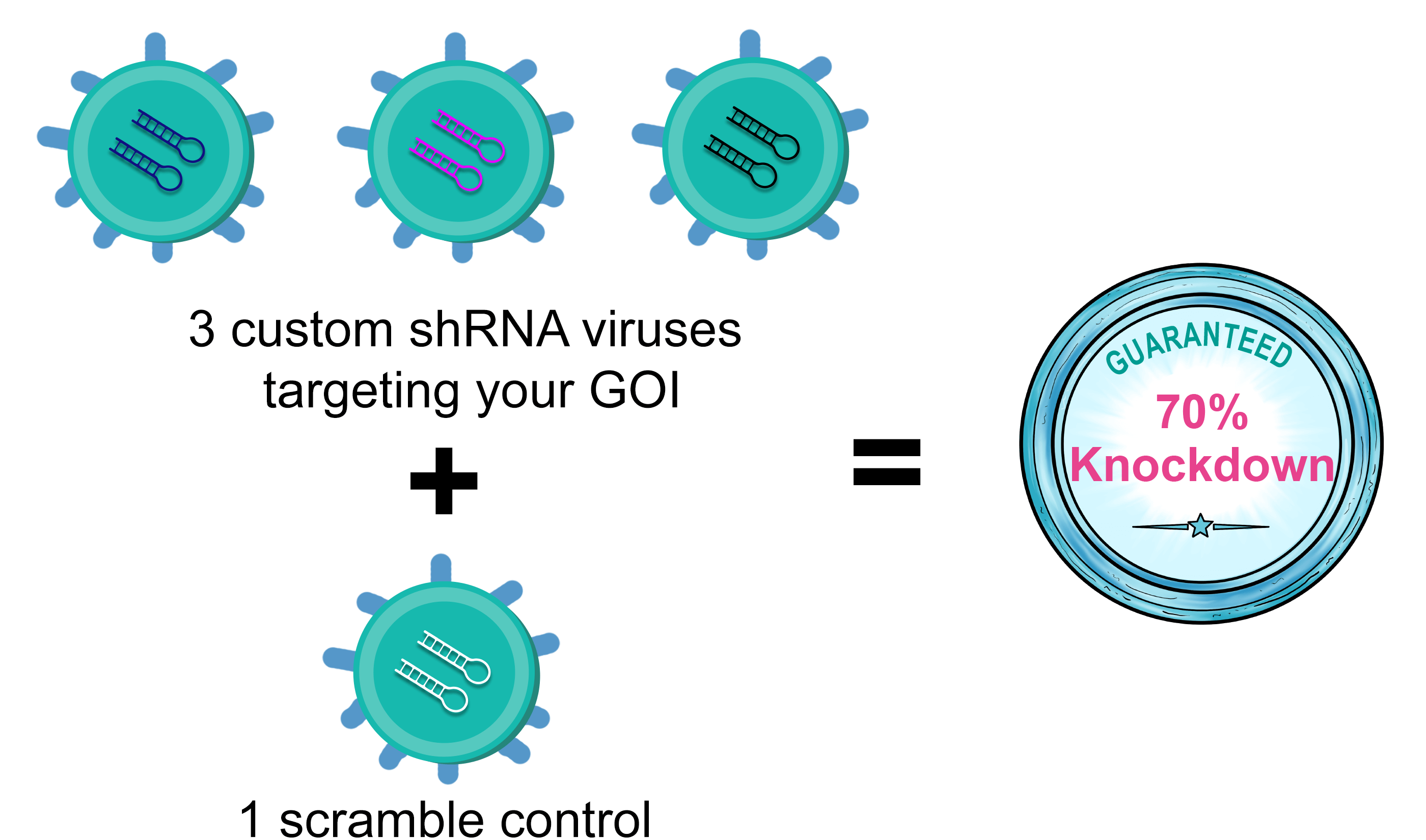
shRNA (3+1) 바이러스 패키징
주문 관련 정보
Price and turnaround Price Match
| Virus Type | Scale & Deliverable | Application | Price (USD)* | Turnaround** |
|---|---|---|---|---|
| Lentivirus | Mini | Cell culture | $899 |
10-19 days
|
| Pilot | $1,499 | |||
| Medium | $1,999 | |||
| Large | $2,999 | |||
| Ultra-purified medium | Cell culture & in vivo | $3,999 | ||
| Ultra-purified large | $4,799 | |||
| AAV | Pilot | Cell culture | $1,499 |
10-19 days
|
| Medium | $1,999 | |||
| Large | $2,999 | |||
| Ultra-purified pilot | Cell culture & in vivo | $4,199 |
11-21 days
|
|
| Ultra-purified medium | $5,699 | |||
| Ultra-purified large | $8,799 | |||
| Adenovirus | Pilot | Cell culture | $2,399 | 34-47 days |
| Medium | $3,599 | |||
| Large | $4,699 | |||
| Ultra-purified medium | Cell culture & in vivo | $6,199 | 34-47 days | |
| Ultra-purified large | $7,499 |
* 가격에는 벡터 제작 및 바이러스 패키징이 모두 포함되어 있습니다.
** 소요시간에는 벡터 제작 및 바이러스 패키징에 대한 생산 시간이 포함됩니다. 최종 결과물을 고객에게 배송하기 위한 운송 시간은 포함되지 않습니다.

Terms and Conditions: VectorBuilder는 shRNA(3+1) 바이러스 패키징 서비스에 대한 knockdown 보증을 제공합니다. VectorBuilder의 추천된 상위 3개로 제작된 shRNA 중 목적유전자(GOI)의 70% 이상 knockdown효율을 달성한 shRNA가 하나도 없을 경우 고객은 서비스 가격의 1/3에 해당하는 스토어크레딧을 받을 수 있습니다. 본 서비스 보증을 받기 위해서는 권장하는 상위 3개의 shRNA를 선택해야 합니다.
아래의 벡터 picker를 사용하여 GOI에 대한 세가지 shRNA를 선택하실 수 있습니다.
위의 벡터 picker는 U6 기반의 shRNA 벡터에만 적용됩니다. miR30 기반의 shRNA 벡터가 필요하거나 벡터 picker를 사용하여 타겟 유전자에 대한 원하는 shRNA를 찾을 수 없는 경우, 원하시는 사항을 디자인 의뢰하기로 보내 주시면 당사의 기술팀이 shRNA 벡터를 디자인해드립니다.
기술적인 정보
제공되는 벡터 시스템
현재 shRNA (3 + 1) 바이러스 패키징 서비스에 사용할 수 있는 바이러스 유형은 렌티바이러스, AAV 및 아데노바이러스입니다. 이러한 모든 벡터 시스템은 E. coli에서 높은 copy 수의 복제, 높은 titer의 살아있는 바이러스 패키징 및 호스트 세포에 효율적인 transduction을 위하여 최적화되어 있습니다. shRNA 발현은 human U6 promoter에 의하며, transduction된 세포 내에서 타겟 유전자 mRNA의 분해를 유도합니다. AAV 패키징의 경우 다음의 serotype을 제공합니다: 1, 2, 3, 4, 5, 6, 6.2, 7, 8, 9, rh10, DJ, DJ/8, PHP.eB, PHP.S, AAV2-retro, AAV2-QuadYF and AAV2.7m8.
자료
자주 묻는 질문
생의학 연구분야에서 많이 사용되는 바이러스 벡터에는 렌티바이러스, 아데노부속바이러스 (AAV), 아데노바이러스가 있으며, 이들은 각각 장단점을 가지고 있습니다. 아래의 표에는 실힘 목적에 적합한 바이러스를 선택할 때 고려해야 하는 중요한 사항들이 요약되어 있습니다.
| Lentivirus | AAV | Adenovirus | |
|---|---|---|---|
| Tropism | Broad | Depending on viral serotype | Ineffective for some cells |
| Can infect non-dividing cells? | Yes | Yes | Yes |
| Stable integration or transient? | Stable integration | Transient, episomal | Transient, episomal |
| Maximum titer | High | High | Very High |
| Promoter customization | Yes | Yes | Yes |
| Primary use | Cell culture and in vivo | In vivo | In vivo |
| Immune response in vivo | Low | Very low | High |
바이러스 입자를 채취한 후에 바이러스 벡터가 형광 reporter 유전자를 가지고 있으면 바이러스의 품질 확인을 위한 첫 단계로 293T 또는 293A와 같은 세포에 transduction하여 형광 단백질의 발현을 확인합니다. Titer를 정량하기 위해서는 바이러스 유형에 따라 각각 다른 방법을 사용합니다. 형광 관측과 정량한 값 사이에 편차기 큰 경우, 제조된 바이러스의 고품질을 보장할 수 있도록 다시 측정을 하거나 추가로 다른 방법으로 검증을 합니다.
렌티바이러스
당사에서는 p24 ELISA에 의하여 렌티바이러스의 titer를 결정합니다. 렌티바이러스가 포함된 세포 배양 상청액에 있는 HIV-1 p24 core 단백질을 정량하기 위하여 sandwich immunoassay를 이용합니다. 먼저 렌티바이러스 샘플 내의 p24와 결합하기 위한 anti-HIV-1 p24 capture antibody로 코팅된 microtiter plate에 상청액을 넣습니다. 그 다음에 biotinylated anti-p24 secondary antibody를 넣으면 p24 antibody에 결합된 p24에 결합을 합니다. Streptavidin-HRP conjugate를 그 다음에 넣으면 streptavidin과 biotin이 결합하며, 최종적으로 HRP와 substrate의 반응에 의하여 발색하게 됩니다. 이 때 나타나는 색의 강도는 렌티바이러스 샘플 내에 존재하는 p24의 양에 비례하며, spectrophotometer를 사용하여 재조합 HIV-1 p24 standard curve와 비교함으로써 정확한 양을 결정할 수 있습니다. p24 수치로부터 최종적으로 렌티바이러스 샘플 내의 바이러스 titer를 얻게 됩니다.
아데노부속바이러스 (AAV)
당사에서는 AAV의 titer를 측정하기 위하여 먼저 용해시킨 바이러스 입자로부터 유전체를 추출합니다. 그 다음에 qPCR 을 사용하여 ITR 서열의 copy 수를 정확히 측정하여 이로부터 바이러스 유전체의 copy 수를 정량합니다. AAV 입자는 매우 안정합니다. 기본적으로 당사의 AAV 제조 방법에 의한 모든 바이러스 입자는 살아 있으며, 상온에서도 며칠 동안 활성을 유지할 수 있습니다. 따라서 transduction에 의한 titer 측정 방법에 의하지 않고 물리적인 방법으로 정해진 titer는 기능적인 titer에 매우 근접합니다.
아데노바이러스
아데노바이러스에 대해서는 기능적인 titer를 측정합니다. 연속적으로 희석한 아데노바이러스를 293A 세포에 transduction한 후에 immunochemistry에 의하여 아데노바이러스의 hexon 단백질을 발현하는 세포를 착색하고, 착색된 세포 하나를 one infectious unit으로 계산합니다. 대부분의 각각의 세포가 하나의 바이러스 입자에 의해서만 transduction이 되도록 매우 낮은 MOI를 사용하며, 이 방법은 기존의 plaque assay와 좋은 상관관계를 보여줍니다. Ultra-purified 아데노바이러스의 경우는 optical density와 기능적인 titer가 높은 상관성을 보여주기 때문에 optical density (OD260) 를 이용하여 titer를 결정합니다. 아데노바이러스는 안정성이 매우 높으며, 기본적으로 모든 바이러스 입자가 살아있으며, 상온에서도 며칠 동안 기능을 유지할 수 있습니다.
VectorBuilder에서는 RNAi consortium (TRC)에서 이용하는 것과 유사한 규칙에 따라서 shRNA의 디자인과 score 계산을 합니다. 하나의 RefSeq transcript에 대하여 candidate target site로 이용될 수 있는 모든 가능한 21mer들을 검색합니다. Candidate들 중에서 knockdown 효율과 특이성 (specificity)을 감소시킬 것으로 생각되거나, 4개 이상의 같은 염기가 반복되거나, 7개 이상의 G 또는 C가 반복되거나, GC 함량이 25% 미만 또는 60%를 초과하거나, 5' end에 AA 서열이 있는 등 클로닝에 문제가 될 수 있는 서열들은 제외됩니다. Candidate가 서열 내부의 stem-loop 구조, 3' end 쪽의 높은 GC 함량, 알려진 miRNA seed 서열을 포함하거나, 다른 유전자에 off-target으로 match되는 경우에는 knockdown score에 불이익을 받게 되어 선택될 가능성이 낮아지게 됩니다. 여러 개의 transcript가 존재하는 유전자의 경우, 모든 transcript에 존재하는 서열이 더 높은 score를 받게 됩니다.
모든 score는 0 이상으로, 평균 5, 표준편차 5 정도의 분포를 나타내며, 95%의 score가 15 이하입니다. Knockdown score가 15 정도인 shRNA들은 최상의 knockdown performance와 클로닝 가능성을 가진 것으로 간주되며, knockdown score가 0 인 경우 최악의 knockdown performance를 가지거나 클로닝이 어려운 shRNA로 간주됩니다.
이러한 knockdown score는 대략적인 가이드에 불과하다는 점에 유의해주시기 바랍니다. 실제 knockdown 효율은 score로 예상되는 것과 큰 차이가 있을 수 있습니다. Score가 낮은 타겟 부위도 실제로는 knockdown을 잘 할 가능성이 있습니다. 타겟 유전자의 3’ UTR도 코딩 부위와 마찬가지로 효과가 있을 수 있습니다.
모든 shRNA가 예상처럼 효과를 나타내지는 않습니다.
당사의 경험과 고객의 피드백에 의하여, 임의의 유전자에 대한 3-4 개의 shRNA를 테스트하는 경우 일반적으로 2-3 개만이 적당하거나 좋은 효과를 나타내는 것을 알고 있습니다. 하지만, shRNA를 사용할 때는 모든 shRNA가 다 효과를 나타내는 것이 아니라는 점을 염두에 두는 것이 중요합니다. 일반적으로 50-70%의 shRNA가 눈에 띄는 효과를 나타내며, 그 중에서 23-30%가 강한 효과를 나타냅니다. 특정한 유전자를 타겟팅하는 몇 개의 shRNA를 사용할 경우, 그 중 하나도 만족스러운 knockdown 효과가 없을 수도 있습니다. 그럴 때 가장 좋은 방법은 문헌을 통하여 검증된 것을 포함하여 몇 가지 shRNA들을 추가로 테스트해보는 것입니다. 많은 연구자들이 서로 다른 shRNA들을 포함하는 “칵테일” 을 사용하기도 하며, 때때로 이에 의하여 knockdown 효율을 향상시킬 수 있음을 보여주고 있습니다.
유전자의 knockdown을 확인하기 위한 분석 방법이 적절하지 않습니다.
shRNA의 knockdown 효율을 평가하기 위한 가장 일반적이고 민감한 분석 방법은 RT-qPCR 입니다. 때때로 여러 쌍의 primer들 중에서 가장 높은 서열 특이성과 PCR 효율을 나타내는 것을 선택해야 할 수도 있습니다. 일반적으로 유전체 DNA를 증폭하는 것을 방지하기 위하여 가능하면 RT-qPCR primer들이 exon-exon junction에 걸쳐 있도록 해야 합니다. 새로운 primer 쌍을 사용할 때에는 PCR 결과물을 agarose gel에서 전기영동을 하여 band를 확인하거나, sequencing에 의하여 확인을 하는 것이 좋습니다. 유전체 DNA에 의한 오염을 더 잘 확인하기 위해서는 minus-RT control을 RT-qPCR을 할 때 포함시켜야 합니다. Primer의 품질을 더 잘 확인하려면 NCBI primer designing tool을 사용해보는 것도 도움이 될 것입니다.
Knockdown 효율은 Western blot에 의해서도 확인될 수 있습니다. 하지만, Western blot에 의하면 비특이적인 항체에 의하여 false positive band가 나타날 수 있음은 이미 잘 알려져 있으며, 이에 의하여 knockdown이 되지 않은 것으로 해석될 수도 있게 합니다. 따라서 사용하는 항체가 GOI에 의해 만들어지는 단백질에 정말로 특이적인 반응을 나타내는 것인지를 확인하는 것이 매우 중요합니다.
shRNA가 원하는 유전자의 transcript isoform들 중에서 일부에만 타겟팅이 되었을 수도 있습니다.
shRNA를 디자인할 때, 특정한 isoform만을 대상으로 할 목적이 아니면 가능한 한 많은 transcript isoform이 타겟이 될 수 있도록 디자인하는 것이 좋습니다. VectorBuilder에서는 일반적으로 많이 이용되는 생물종에 대한 최적화된 shRNA들을 포함하는 데이터베이스를 만들었습니다. VectorBuilder 온라인 플랫폼에서 shRNA 벡터를 디자인하면서 shRNA 구성 요소를 지정할 때 데이터베이스 내에서 원하는 유전자로 검색을 할 수 있습니다. 그 다음에 shRNA 들에 대한 상세한 정보를 볼 수 있으며, 이 때 UCSC Genome Browser link를 통하여 유전체 서열과 transcript isoform 상에서 shRNA를 볼 수 있습니다.


