Me First! How Do I Optimize a Vector with Multiple Genes?
Gene delivery systems allow for efficient introduction of not just one gene, but often multiple components, including a promoter, open reading frame (ORF), and marker. These systems can also be customized to meet individual experimental needs by creating vectors with multiple ORFs, multiple fused or linked reporters, and even full expression cassettes. Efficient delivery of these vectors requires careful consideration of many points, especially when using lentiviral vectors. VectorBuilder is here to guide you through key factors related to design and optimization of vectors with multiple ORFs.
Different genes, different drivers
One option for introducing multiple genes is to insert separate expression cassettes, particularly if utilizing different promoters or systems like piggyBac with large carrying capacity. In this instance, an individual promoter, ORF, and polyA tail are inserted for each gene of interest to ensure separate transcription events. This is commonly seen when designing all-in-one inducible gene expression vectors, which contain (1) the gene of interest driven by the TRE promoter and (2) the Tet regulator protein(s) driven by one or more promoters.
Importantly, while designing multiple expression cassettes is possible in most vector systems, it is not suitable in lentivirus. The long terminal repeats (LTRs) in lentivirus both facilitate genomic integration and mark the start and stop of transcription of the recombinant viral genome. As the 3' LTR acts as a polyadenylation signal, including an internal polyA in the lentiviral vector will prematurely terminate transcription. The 3' LTR will therefore not be transcribed, leading to a decrease in viral titer.
To restart or to skip
For lentivirus, and commonly for vector systems, a popular option for introducing multiple ORFs is by creating a polycistron or multicistron, where multiple genes are placed downstream of a single promoter with linkers in between. All of the ORFs are transcribed together as a single mRNA strand, with linkers facilitating translation of separate proteins. The most commonly used linkers are IRES and 2A peptides (P2A, T2A, E2A, F2A). These two classes are compared in the table below.
| Linker Type | Mechanism | Advantage | Disadvantage |
|---|---|---|---|
| IRES | Additional ribosomal recruitment site, initiating translation at an internal site within mRNA | ||
| 2A Peptide | Self-cleaving peptides that cause the ribosome to “skip” synthesis of a peptide bond within the linker |
Considering these factors, we recommend selecting 2A peptides when using more than two ORFs, or if near-equivalent expression levels of the ORFs are required. Of note, when expressing more than two ORFs, different linkers should be used (e.g. P2A and T2A) to avoid recombination. Within the 2A peptide family we recommend P2A, as it has the highest cleavage efficiency, or T2A, which has slightly lower efficiency. Use of IRES is recommended when lower expression levels of the second ORF do not affect the experimental outcome, for instance when the second ORF is a drug-selection marker.
Do the shuffle
After deciding how to separate the chosen ORFs, some further considerations can aid in optimization, particularly the order in which ORFs are placed. When optimizing polycistronic expression, an intriguing and seldom studied question persists: what is the influence of relative positioning on expression profiles?
Previous studies using multiple fluorescent proteins have shown that the expression level of each gene depends on its respective position within the polycistronic transcript. To examine the effects of different arrangements on expression profiles in a more physiologically relevant context, we compared expression of three human endogenous genes. They are expressed from six polycistronic lentiviral vectors containing the full factorial arrangement of the three genes. Intervening P2A and T2A were used as the first and second linker, respectively (Fig. 1A).
Upon comparing the expression levels of a specific gene in different positions (Fig. 1B and 1C), highest expression levels were typically found when the ORF was placed closest to the promoter. The lowest expression was consistently observed when the ORF was placed in the third position, with a reduction of at least 80% compared to the first position.
Intriguingly, relative expression levels of an individual gene can be influenced by not only its own position, but also by the sequential order of the other two genes (Fig. 1B). For instance, expression levels of Gene B were highest when it was placed in first position (Vectors 3 and 4); however expression of Gene B increased 5-fold when Gene C was in second position compared to Gene A (Vector 3 vs Vector 4, respectively). In both vectors, expression level of the second gene was also dramatically affected.
While the expression of a particular ORF can decline dramatically as its position moves toward the end of the construct, expression is also affected by the arrangement of other ORFs within the polycistronic construct. By optimizing the position of your chosen genes, you can achieve experiment-specific expression goals.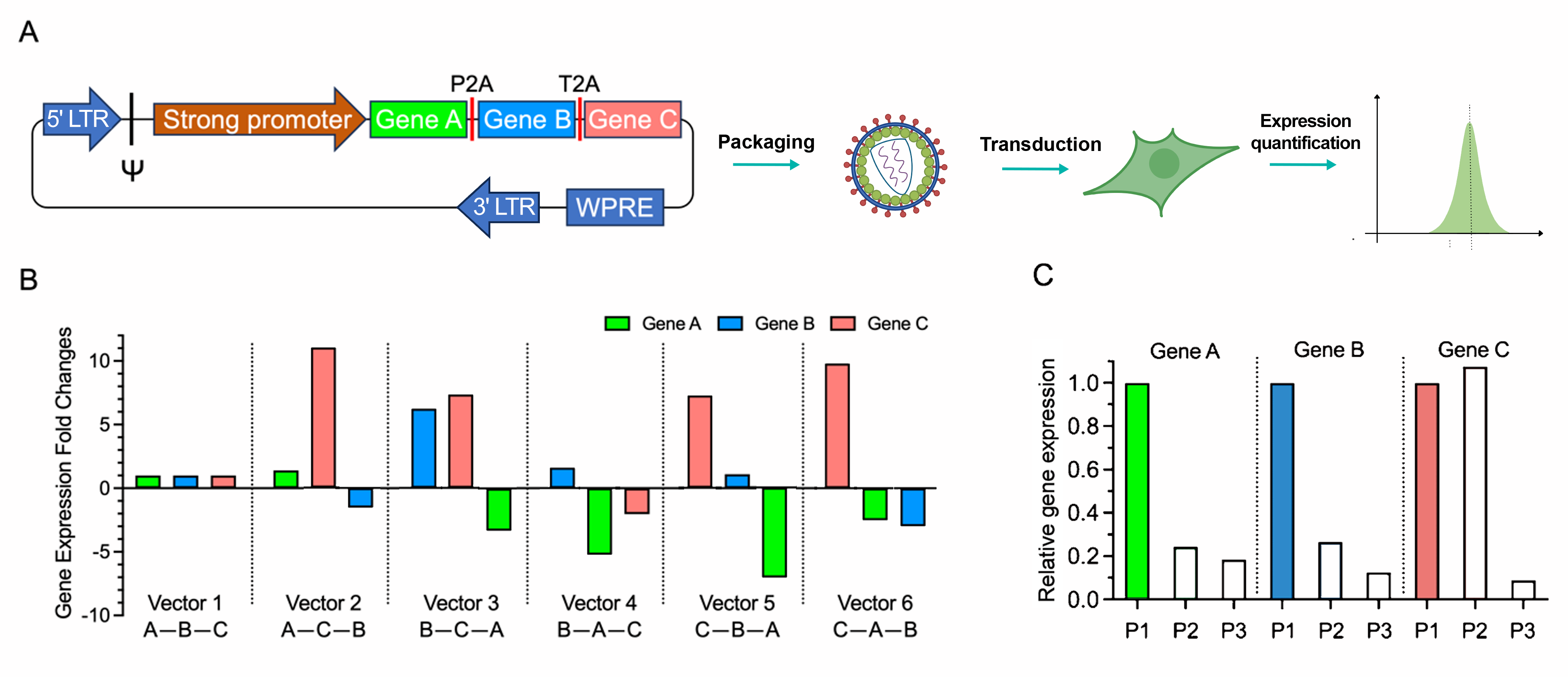 Figure 1. Comparison of position-dependent gene expression levels in 2A linked polycistronic vectors. (A) A tri-cistronic lentiviral vector carrying three ORFs with intervening P2A and T2A peptides was packaged into lentivirus. Mammalian cells were then transduced, and the expression of each gene was evaluated across all six arrangements of the three ORFs by flow cytometry. (B) The six arrangements included A-B-C, A-C-B, B-C-A, B-A-C, C-B-A and C-A-B. To facilitate more straightforward comparisons, we normalized the expression level of each gene within any given arrangement relative to its expression level in the A-B-C order. (C) Protein expression levels at specific positions within the tri-cistronic vector were compared. We normalized the expression level of each gene in different positions to its expression level in the first position.
Figure 1. Comparison of position-dependent gene expression levels in 2A linked polycistronic vectors. (A) A tri-cistronic lentiviral vector carrying three ORFs with intervening P2A and T2A peptides was packaged into lentivirus. Mammalian cells were then transduced, and the expression of each gene was evaluated across all six arrangements of the three ORFs by flow cytometry. (B) The six arrangements included A-B-C, A-C-B, B-C-A, B-A-C, C-B-A and C-A-B. To facilitate more straightforward comparisons, we normalized the expression level of each gene within any given arrangement relative to its expression level in the A-B-C order. (C) Protein expression levels at specific positions within the tri-cistronic vector were compared. We normalized the expression level of each gene in different positions to its expression level in the first position.
Conclusion
Multiple genes can be expressed in target cells through a variety of methods, dependent on the cell type, vector delivery system, number of promoters required, and the specific expression requirements of the different genes. When using lentiviral vectors, there are notable limitations, as separate expression cassettes cannot be used on a single vector. However, by carefully considering linkers and examining order of your genes of interest, expression levels can be optimized for your experiment. VectorBuilder’s technical support is always on-hand to help with this optimization, taking advantage of our decades of collective experience in cloning and packaging complex vectors.
Source
Guntaka RV. Transcription termination and polyadenylation in retroviruses. Microbiol Rev. 1993 Sep;57(3):511-21. doi: 10.1128/mr.57.3.511-521.1993. PMID: 7902524; PMCID: PMC372924.
Hager S, Frame FM, Collins AT, Burns JE, Maitland NJ. An internal polyadenylation signal substantially increases expression levels of lentivirus-delivered transgenes but has the potential to reduce viral titer in a promoter-dependent manner. Hum Gene Ther. 2008 Aug;19(8):840-50. doi: 10.1089/hum.2007.165. PMID: 18627247.
Liu Z, Chen O, Wall JBJ, Zheng M, Zhou Y, Wang L, Vaseghi HR, Qian L, Liu J. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci Rep. 2017 May 19;7(1):2193. doi: 10.1038/s41598-017-02460-2. PMID: 28526819; PMCID: PMC5438344.
Shaimardanova AA, Kitaeva KV, Abdrakhmanova II, Chernov VM, Rutland CS, Rizvanov AA, Chulpanova DS, Solovyeva VV. Production and Application of Multicistronic Constructs for Various Human Disease Therapies. Pharmaceutics. 2019 Nov 6;11(11):580. doi: 10.3390/pharmaceutics11110580. PMID: 31698727; PMCID: PMC6920891.



