Vector Systems
CRISPR Interference (dCas9-KRAB-Mediated) Lentiviral Vector
Overview
CRISPR/Cas9 vectors are among several types of emerging genome editing tools that can quickly and efficiently create mutations at target sites of a genome (the other two popular ones being ZFN and TALEN).
Cas9 is a member of a class of RNA-guided DNA nucleases which are part of a natural prokaryotic immune system that confers resistance to foreign genetic elements such as plasmids and bacteriophage. Within the cell, the Cas9 enzyme forms a complex with a guide RNA (gRNA), which provides targeting specificity through direct interaction with homologous 18-22nt target sequences in the genome. Hybridization of the gRNA to the target site localizes Cas9, which then cuts the target site in the genome.
The dCas9-KRAB system is a powerful tool for transcriptional repression of genes within their endogenous genomic loci. This system is derived from CRISPR/Cas9 genome-editing systems, but rather than mediating genome editing, a catalytically inactive form of the Cas9 protein (dCas9) is utilized to direct the assembly of a transcriptional repression complex at target sites in the genome. While dCas9 still retains the ability to bind to DNA target sites when used in conjunction with a targeting gRNA, it can no longer cleave target DNA sequences.
The complete dCas9-KRAB system consists of two components, each provided in separate lentiviral vectors. The first component is the gRNA expression vector driving the expression of the gRNA specific to the DNA target site of interest which is usually the promoter region of the target gene. The second component required for this system is the dCas9-KRAB helper vector which drives the expression of a fusion protein consisting of dCas9, a catalytically inactive variant of Cas9 and the transcriptional repressor KRAB (Kruppel-associated box domain). Two versions of the dCas9-KRAB helper vector are currently available – the original dCas9-KRAB helper vector version and an improved dCas9-KRAB-MeCP2 helper vector version which drives the expression of dCas9 fused to a bipartite repressor domain, KRAB-MeCP2 for achieving more potent transcriptional repression of DNA target sites.
The gRNA expression vector can be used either with the dCas9-KRAB helper vector or the dCas9-KRAB-MeCP2 helper vector for achieving transcriptional repression of DNA target sites of interest. When cells are co-transduced with these two vectors, the user-selected gRNA recruits either dCas9-KRAB or dCas9-KRAB-MeCP2 via CRISPR-Cas9 complex assembly to gRNA target sites, thereby resulting in their transcriptional inhibition.
Our lentiviral gRNA expression vector for the dCas9-KRAB system is available for expressing either single-gRNA or dual-gRNAs. While single-gRNA vectors are suitable for conventional CRISPRi applications of achieving transcriptional repression of a single genomic target site, dual-gRNA vectors can be used to achieve combinatorial repression of two different genomic target sites of interest simultaneously, for studying genetic interactions. Alternatively, dual-gRNA vectors can also be used for targeting the same genomic region with two different gRNAs to achieve stronger levels of repression compared to that obtained with a single gRNA. While the single-gRNA vector consists of a single human U6 promoter driving the target site-specific gRNA sequence, the dual gRNA vector consists of two consecutive U6 promoters driving the expression of two gRNA sequences specific to either the same genomic target site or two different genomic sites of interest.
For further information about this vector system, please refer to the papers below.
| References | Topic |
|---|---|
| Cell. 154:442 (2013) | Characterization of CRISPRa and CRISPRi systems |
| Nat Methods. 12:1143 (2015) | Characterization of the dCas9-KRAB system |
| Nat Methods. 15:611 (2018) | Dual-gRNA and dCas9-KRAB-MeCP2 based gene repression |
Highlights
Our lentiviral dCas9-KRAB vectors are derived from the third-generation lentiviral vector system. This system is optimized for high copy number replication in E. coli, high-titer packaging of live virus, efficient viral transduction of a wide range of cells, and efficient vector integration into the host genome. The human U6 promoter drives high-level, constitutive transcription of the user-selected gRNA sequence to target the genomic DNA site of interest.
Experimental Validation
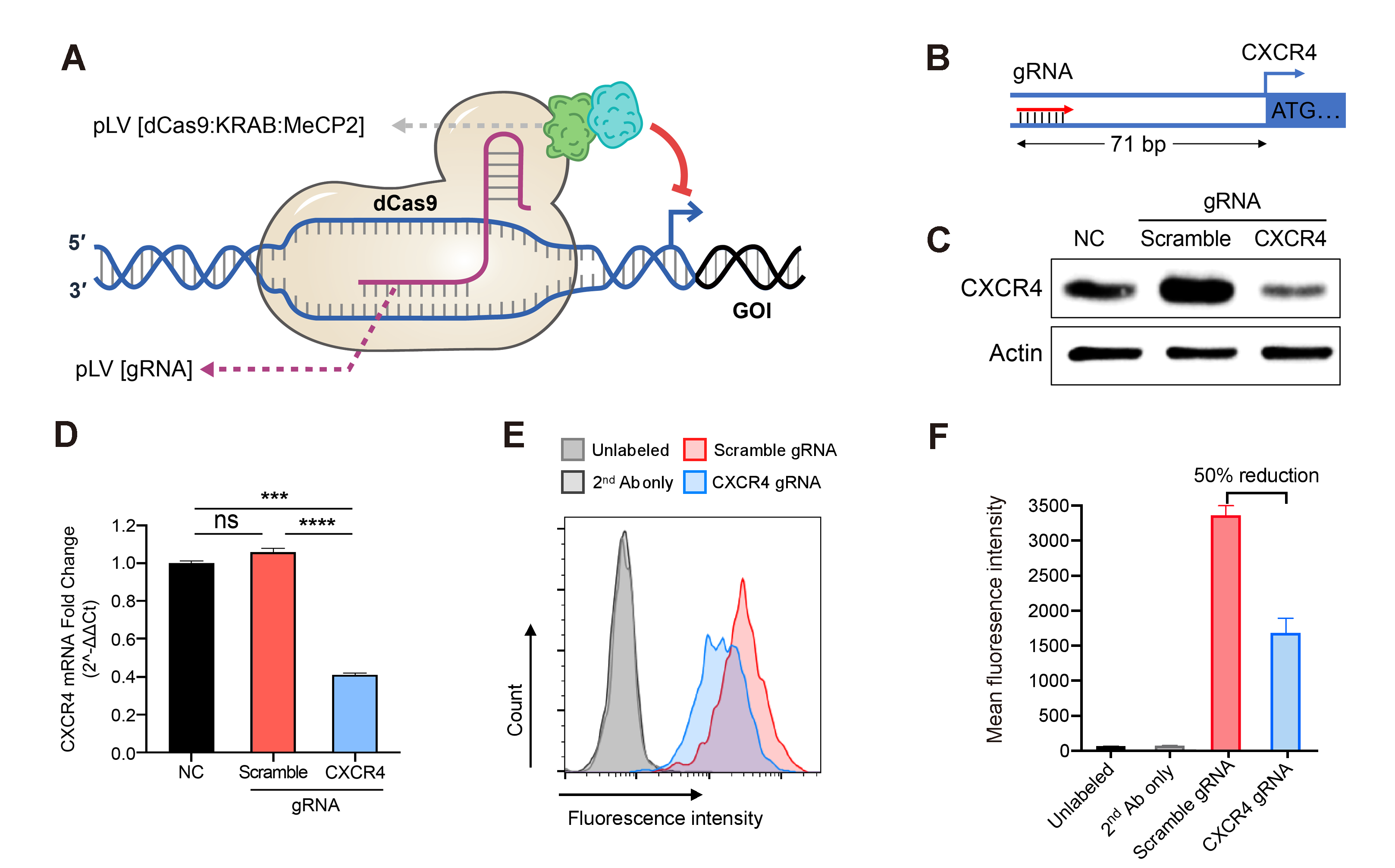
Figure 1. Down-regulation of gene expression achieved by the lentivirus-based CRISPRi. Jurkat cells stably expressing the dCas9/KRAB/MeCP2 transcriptional repressor complex were transduced with gRNA expression lentivirus followed by antibiotic selection. (A) Illustration of dCas9/KRAB/MeCP2 regulated gene transcriptional inhibition. (B) Diagram of gRNA design targeting the promoter region of the human CXCR4 gene. (C) CXCR4 protein levels in Jurkat cells transduced with scramble or targeting gRNA or no treatment control (NC), measured by western blot. (D) Relative CXCR4 gene expression in Jurkat cells transduced with scramble or targeting gRNA or no treatment control (NC), measured by qRT-PCR. Mean±SD, ***P<0.001, ****P<0.0001, ANOVA with Tukey’s post hoc test. (E) The surface expressed CXCR4 in the Jurkat cells transduced with scramble or targeting gRNA were quantified by flowcytometry. CXCR4 was labeled with monoclonal primary antibodies (Ab) and fluorophore-conjugated secondary Ab. Unlabeled and secondary Ab. only Jurkat cells were used as negative controls. (F) The amount of CXCR4 on the surface of cells transduced with the CXCR4 targeting gRNA was averagely reduced by about 50% compared to the cells transduced with the scramble gRNA. Mean±SD.
Advantages
Endogenous genomic context: The dCas9-KRAB system can repress transcription of target sites within their endogenous genomic loci. This is unlike transgenic or genome-editing methods which involve alterations to the genomic context of the gene of interest.
Strong repression: Transcriptional repression of genes using the dCas9-KRAB system can often achieve very high-level gene repression.
Broad coverage: Since the dCas9-KRAB system represses target sites at the DNA level, it can be used to target a variety of transcripts including non-coding RNAs, microRNAs, antisense transcripts, nuclear localized RNAs, and polymerase III transcripts.
Specificity: The dCas9-KRAB system can achieve highly specific repression of DNA target sites with minimal off-target effects.
Disadvantages
Technical complexity: The use of lentiviral vectors requires the production of live virus in packaging cells followed by the measurement of viral titer. These procedures are technically demanding and time consuming relative to conventional plasmid transfection.
Requires multiple vectors: This vector system requires co-expression of either dCas9-KRAB or dCas9-KRAB-MeCP2 with gRNA, and separate vectors should be used for these components.
Gene-by-gene variation: The extent of transcriptional repression achieved with the dCas9-KRAB system might vary from one gene to another depending upon their endogenous chromatin states.
Key components
Single-gRNA expression lentiviral vector
RSV promoter: Rous sarcoma virus promoter. It drives transcription of viral RNA in packaging cells. This RNA is then packaged into live virus.
5' LTR-ΔU3: A deleted version of the HIV-1 5' long terminal repeat. In wildtype lentivirus, 5' LTR and 3' LTR are essentially identical in sequence. They reside on two ends of the viral genome and point in the same direction. Upon viral integration, the 3' LTR sequence is copied onto the 5' LTR. The LTRs carry both promoter and polyadenylation function, such that in wildtype virus, the 5' LTR acts as a promoter to drive the transcription of the viral genome, while the 3' LTR acts as a polyadenylation signal to terminate the upstream transcript. On our vector, 5' LTR-ΔU3 is deleted for a region that is required for the LTR's promoter activity normally facilitated by the viral transcription factor Tat. This does not affect the production of viral RNA during packaging because the promoter function is supplemented by the RSV promoter engineered upstream of 5'LTR-ΔU3 LTR.
Ψ: HIV-1 packaging signal required for the packaging of viral RNA into virus.
RRE: HIV-1 Rev response element. It allows the nuclear export of viral RNA by the viral Rev protein during viral packaging.
cPPT: HIV-1 Central polypurine tract. It creates a "DNA flap" that increases nuclear import of the viral genome during target cell infection. This improves vector integration into the host genome, resulting in higher transduction efficiency.
U6 promoter: Drives expression of the user-selected downstream gRNA sequence. This is the promoter of the human U6 snRNA gene, an RNA polymerase III promoter which efficiently expresses short RNAs.
gRNA: Guide RNA compatible with Cas9 derived from Streptococcus pyogenes.
Terminator: Terminates transcription of the gRNA.
hPGK promoter: Human phosphoglycerate kinase 1 gene promoter. It drives the ubiquitous expression the downstream marker gene.
Marker: A drug selection gene (such as neomycin resistance), a visually detectable gene (such as EGFP), or a dual-reporter gene (such as EGFP/Neo). This allows cells transduced with the vector to be selected and/or visualized.
WPRE: Woodchuck hepatitis virus posttranscriptional regulatory element. It enhances transcriptional termination in the 3' LTR during viral RNA transcription, which leads to higher levels of functional viral RNA in packaging cells and hence greater viral titer. It also enhances transcriptional termination during the transcription of the user's gene of interest on the vector, leading to their higher expression levels.
ΔU3/3' LTR: A truncated version of the HIV-1 3' long terminal repeat that deletes the U3 region. This leads to the self-inactivation of the promoter activity of the 5' LTR upon viral vector integration into the host genome (since the 3' LTR is copied onto 5' LTR during viral integration). The polyadenylation signal contained in 3' LTR-ΔU3 serves to terminates all upstream transcripts produced both during viral packaging and after viral integration into the host genome.
SV40 early pA: Simian virus 40 early polyadenylation signal. It further facilitates transcriptional termination after the 3' LTR during viral RNA transcription during packaging. This elevates the level of functional viral RNA in packaging cells, thus improving viral titer.
Ampicillin: Ampicillin resistance gene. It allows the plasmid to be maintained by ampicillin selection in E. coli.
pUC ori: pUC origin of replication. Plasmids carrying this origin exist in high copy numbers in E. coli.
Dual-gRNA gRNA expression lentiviral vector
RSV promoter: Rous sarcoma virus promoter. It drives transcription of viral RNA in packaging cells. This RNA is then packaged into live virus.
5' LTR-ΔU3: A deleted version of the HIV-1 5' long terminal repeat. In wildtype lentivirus, 5' LTR and 3' LTR are essentially identical in sequence. They reside on two ends of the viral genome and point in the same direction. Upon viral integration, the 3' LTR sequence is copied onto the 5' LTR. The LTRs carry both promoter and polyadenylation function, such that in wildtype virus, the 5' LTR acts as a promoter to drive the transcription of the viral genome, while the 3' LTR acts as a polyadenylation signal to terminate the upstream transcript. On our vector, Δ5' LTR is deleted for a region that is required for the LTR's promoter activity normally facilitated by the viral transcription factor Tat. This does not affect the production of viral RNA during packaging because the promoter function is supplemented by the RSV promoter engineered upstream of Δ5' LTR.
Ψ: HIV-1 packaging signal required for the packaging of viral RNA into virus.
RRE: HIV-1 Rev response element. It allows the nuclear export of viral RNA by the viral Rev protein during viral packaging.
cPPT: HIV-1 Central polypurine tract. It creates a "DNA flap" that increases nuclear importation of the viral genome during target cell infection. This improves vector integration into the host genome, resulting in higher transduction efficiency.
U6 Promoter: Drives expression of the user-selected downstream gRNA sequence. This is the promoter of the human U6 snRNA gene, an RNA polymerase III promoter which efficiently expresses short RNAs.
gRNA #1: The first guide RNA compatible with Cas9 derived from Streptococcus pyogenes.
gRNA #2: The second guide RNA compatible with Cas9 derived from Streptococcus pyogenes.
Terminator: Terminates transcription of the gRNA.
hPGK promoter: Human phosphoglycerate kinase 1 promoter. It drives the ubiquitous expression of the downstream marker gene.
Marker: A drug selection gene (such as neomycin resistance), a visually detectable gene (such as EGFP), or a dual-reporter gene (such as EGFP/Neo). This allows cells transduced with the vector to be selected and/or visualized.
WPRE: Woodchuck hepatitis virus posttranscriptional regulatory element. It enhances viral RNA stability in packaging cells, leading to higher titer of packaged virus.
3' LTR-ΔU3: A truncated version of the HIV-1 3' long terminal repeat that deletes the U3 region. This leads to the self-inactivation of the promoter activity of the 5' LTR upon viral vector integration into the host genome (since 3' LTR is copied onto 5' LTR during viral integration). The polyadenylation signal contained in ΔU3/3' LTR serves to terminates all upstream transcripts produced both during viral packaging and after viral integration into the host genome.
SV40 early pA: Simian virus 40 early polyadenylation signal. It further facilitates transcriptional termination after the 3' LTR during viral RNA transcription during packaging. This elevates the level of functional viral RNA in packaging cells, thus improving viral titer.
Ampicillin: Ampicillin resistance gene. It allows the plasmid to be maintained by ampicillin selection in E. coli.
pUC ori: pUC origin of replication. Plasmids carrying this origin exist in high copy numbers in E. coli.



